Clusters Update¶
It is possible to update a hierarchy of clusters by adding new data after a clustering run was concluded.
Similarly to a k-NN run, this step requires the dataset to be added,
the original dataset used to build the clusters, their membership
table (as output by cluster)
and the path to the reference folder (raccoon_data)
containing the trained maps. It also takes an output folder, a debugging mode switch and a GPU switch.
As in the case of classification and clustering, a wrapper function is available. This will set up the necessary folders and run the update automatically.
import raccoon as rc
updated_membership = rc.update(new_df, original_df, cluster_membership, tolerance=1e-2,
prob_cut=.2, refpath=r'original_clusters/raccoon_data',
dim=2, filter_feat='variance', optimizer='auto',
metric_clu='euclidean', metric_map='cosine',
dyn_mesh=True, chk=True, out_path=r'./',
max_depth=-1,save_map=True, debug=True,
gpu=gpu)
This tool will first project the new samples on the old hierarchy, identify their closest
matching classes with k-NN and re-evaluate their clustering score including the new data.
Iteratively, if the score degrades beyond the given tolerance the clusters
and those along the subsequent branch will be rebuild from scratch, but including the new data.
The new classes are marked with a ‘u’ suffix to the original name at the point of rebuilding.
A second flag, prob_cut, defines a probability cutoff when assigning the best matching
class at each level. Probabilities below this cutoff will be ignored, if a point doesn’t reach
any this threshold with any class it will be considered as noise.
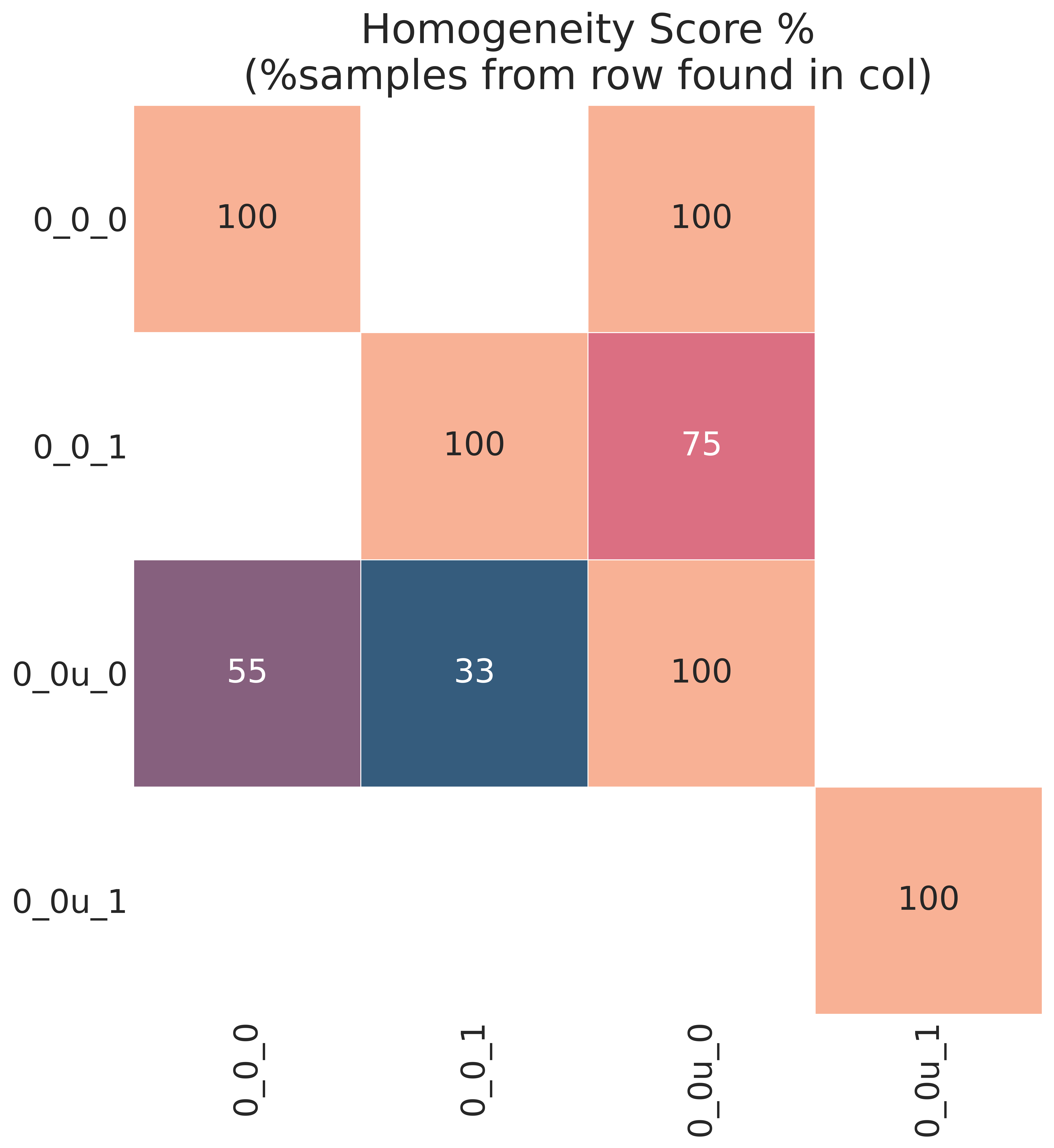
A homogeneity score heatmap showing overlap between the old and new classes,
calculated on the old data only, is also produced in the raccoon_plot folder.
Keywords arguments can be provided for the re-clustering step. These should ideally match the
original setup but don’t have to. If you want to extend the search in this specific instance,
for example, it may be worth changing some of these flags (e.g. max_depth or pop_cut).
Alternatively, the update object can be initialized and the process can be called directly.
from raccoon.utils.update import UpdateClusters
obj = UpdateClusters(new_data, old_data, membership, refpath=r'./raccoon_data',
out_path=out_path, tolerance=1e-2, prob_cut=.2, **kwargs)
obj.find_and_update()
output = obj.new_clus
The output is in the same one-hot-encoded matrix format (rows as samples, columns classes) as the iterative clustering output table.